Motifs detected by DREME on the MeRIP-seq datasets
Par un écrivain mystérieux
Last updated 03 juin 2024

Download scientific diagram | Motifs detected by DREME on the MeRIP-seq datasets from publication: A novel algorithm for calling mRNA m 6 A peaks by modeling biological variances in MeRIP-seq data | Motivation: N⁶-methyl-adenosine (m⁶A) is the most prevalent mRNA methylation but precise prediction of its mRNA location is important for understanding its function. A recent sequencing technology, known as Methylated RNA Immunoprecipitation Sequencing technology (MeRIP-seq), | mRNA, RNA Sequence Analysis and Base Sequence | ResearchGate, the professional network for scientists.
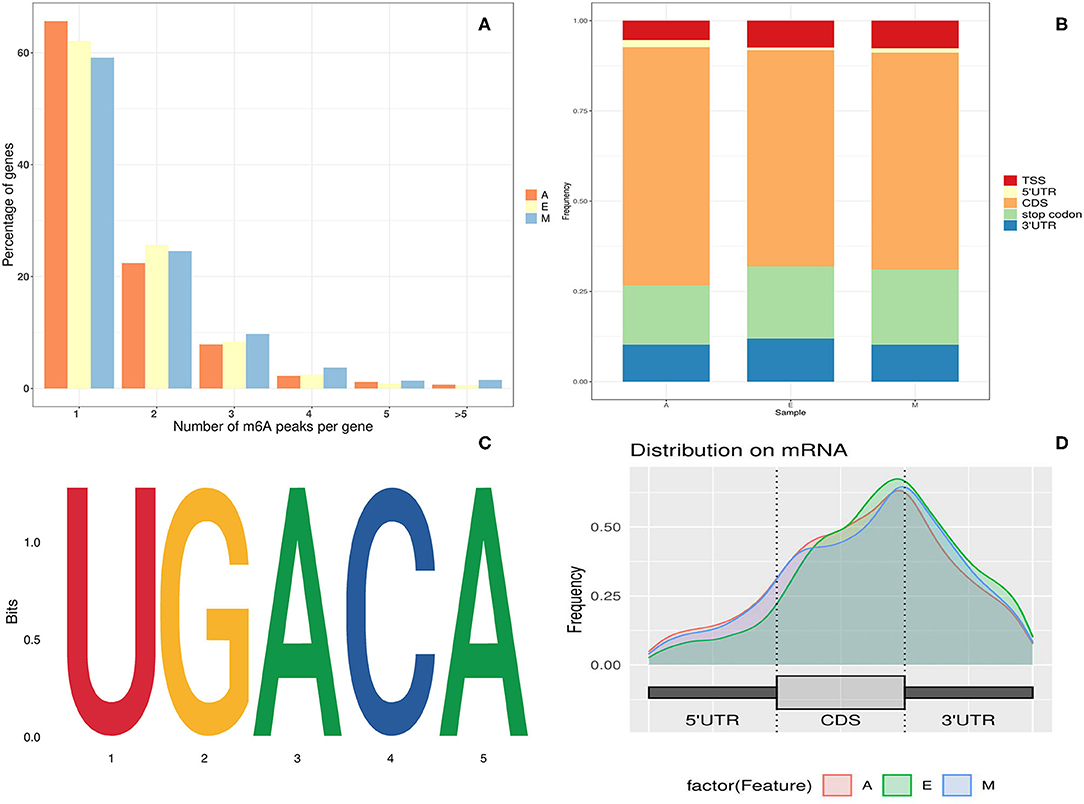
Frontiers Regulatory Role of N6-Methyladenosine in Longissimus Dorsi Development in Yak
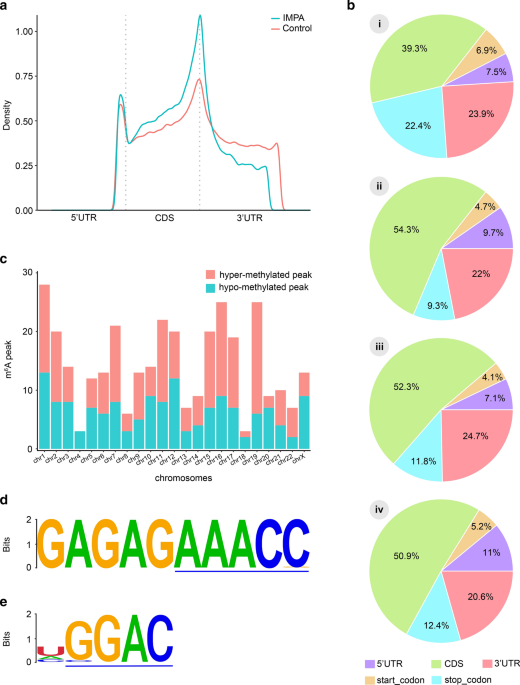
Comprehensive analysis of the transcriptome‐wide m6A methylome in invasive malignant pleomorphic adenoma, Cancer Cell International
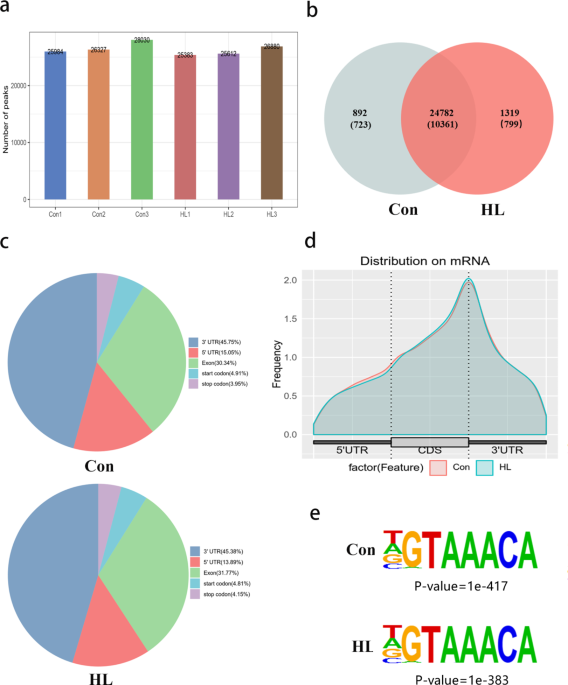
Comprehensive analysis of N6-methyladenosine-related RNA methylation in the mouse hippocampus after acquired hearing loss, BMC Genomics

Comprehensive analysis of transcriptomic profiling of 5‐methylcytosin modification in placentas from preeclampsia and normotensive pregnancies - Wei - 2023 - The FASEB Journal - Wiley Online Library

A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package - ScienceDirect

Refined RIP-seq protocol for epitranscriptome analysis with low input materials. - Abstract - Europe PMC
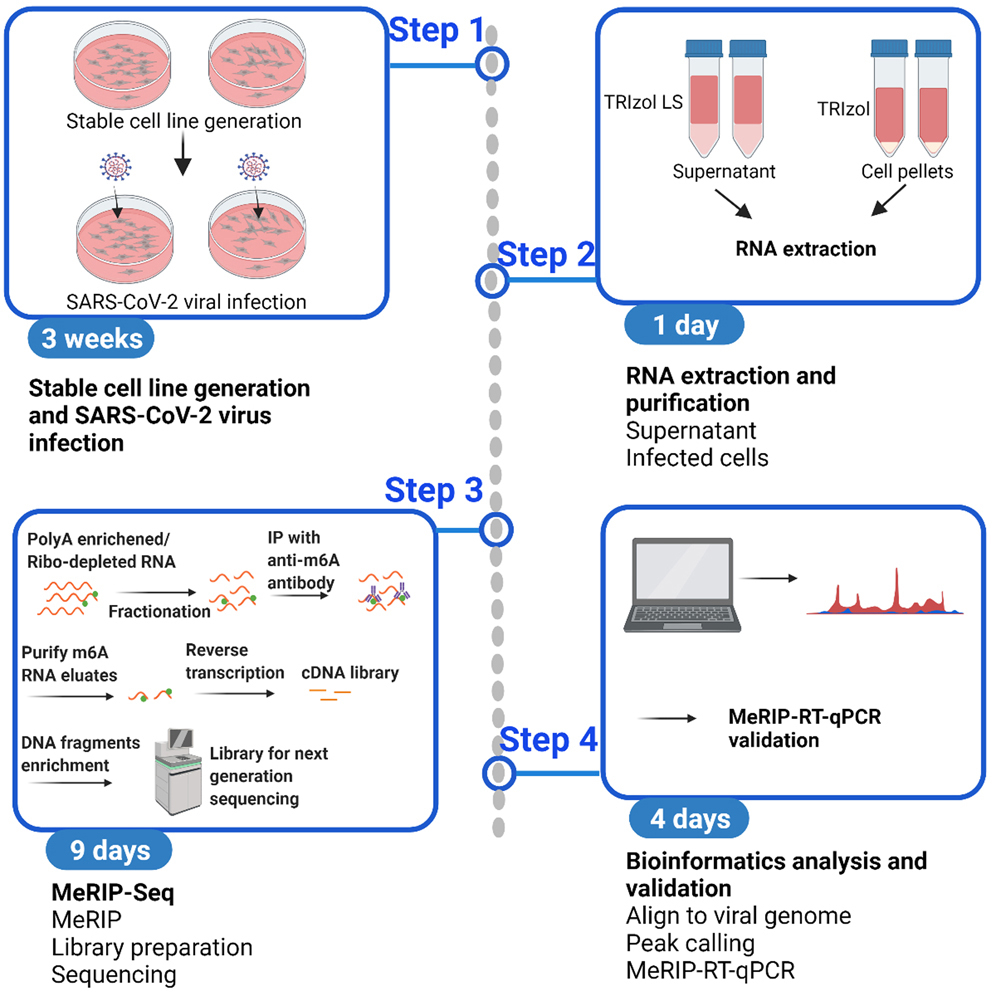
Detection of N6-methyladenosine in SARS-CoV-2 RNA by methylated RNA immunoprecipitation sequencing

A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package - ScienceDirect

Limits in the detection of m6A changes using MeRIP/m6A-seq

DENA: training an authentic neural network model using Nanopore sequencing data of Arabidopsis transcripts for detection and quantification of N6-methyladenosine on RNA
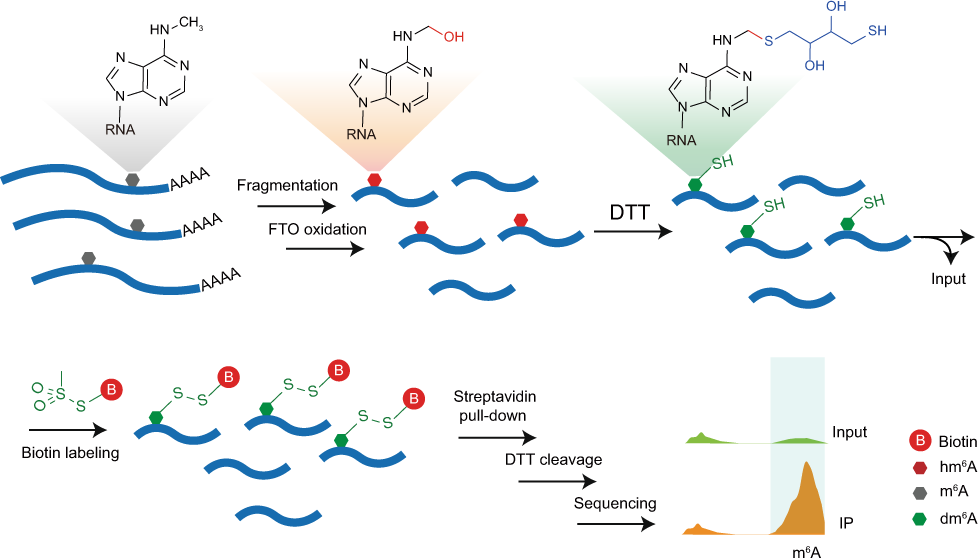
Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine
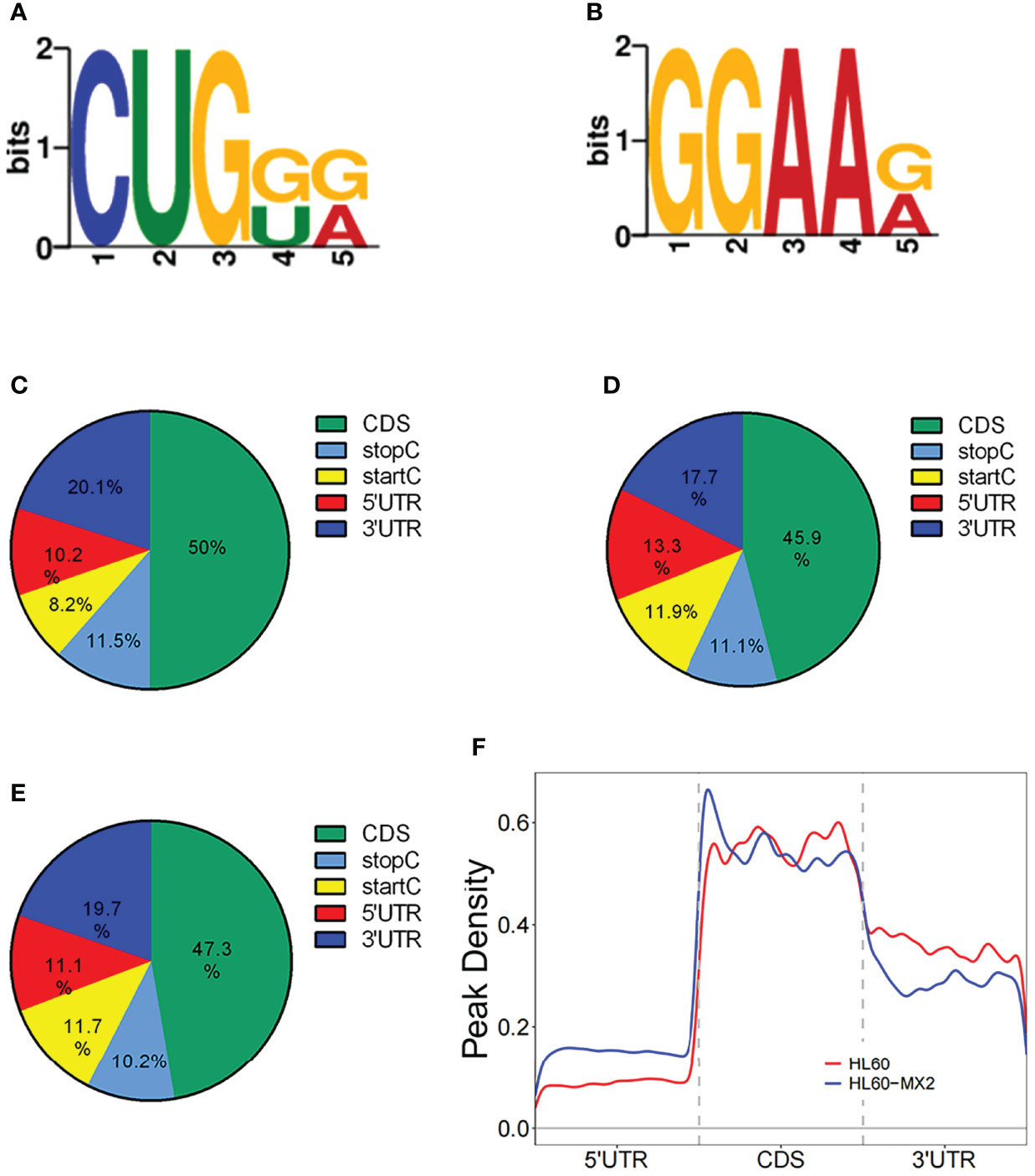
Frontiers Transcriptome Profiling of N7-Methylguanosine Modification of Messenger RNA in Drug-Resistant Acute Myeloid Leukemia

Motifs detected by DREME on the MeRIP-seq datasets
Recommandé pour vous
 100 PIECE Accessories Dreme l Set Variable Speed Rotary Cutter Tool Kit Grinder14 Jul 2023
100 PIECE Accessories Dreme l Set Variable Speed Rotary Cutter Tool Kit Grinder14 Jul 2023 About DREME TE14 Jul 2023
About DREME TE14 Jul 2023 Dreme, Sleepers, Recliners & Gliders14 Jul 2023
Dreme, Sleepers, Recliners & Gliders14 Jul 2023 DREME Foundation (@DREMEfoundation) / X14 Jul 2023
DREME Foundation (@DREMEfoundation) / X14 Jul 2023- Barberz Dreme Co14 Jul 2023
 Hitman Novel14 Jul 2023
Hitman Novel14 Jul 2023 DREME by Samey and Rytmus on Beatsource14 Jul 2023
DREME by Samey and Rytmus on Beatsource14 Jul 2023 Shop Dremel 4000 Multipurpose Rotary Tool Collection at14 Jul 2023
Shop Dremel 4000 Multipurpose Rotary Tool Collection at14 Jul 2023 EU - 150pcs - Graveur Mini Perceuse Électrique 180w, Outils Électriques, Outil Rotatif, Polisseuse Avec Dreme - Cdiscount Bricolage14 Jul 2023
EU - 150pcs - Graveur Mini Perceuse Électrique 180w, Outils Électriques, Outil Rotatif, Polisseuse Avec Dreme - Cdiscount Bricolage14 Jul 2023 Development and Research in Early Mathematics Education (DREME) (DREMEmath) - Profile14 Jul 2023
Development and Research in Early Mathematics Education (DREME) (DREMEmath) - Profile14 Jul 2023
Tu pourrais aussi aimer
 Intégration nouveaux XIAOMI Mijia thermomètre Bluetooth 2 (carrés) dans Blea - Communication - Communauté Jeedom14 Jul 2023
Intégration nouveaux XIAOMI Mijia thermomètre Bluetooth 2 (carrés) dans Blea - Communication - Communauté Jeedom14 Jul 2023 Veste d'hiver Marvel Spider-man bleu - Veste d'hiver pour enfants - Veste pour enfants14 Jul 2023
Veste d'hiver Marvel Spider-man bleu - Veste d'hiver pour enfants - Veste pour enfants14 Jul 2023 Valiant Double-Sided Foam Tape 18mmX1m14 Jul 2023
Valiant Double-Sided Foam Tape 18mmX1m14 Jul 2023 BOLLY - Tapis Tissé 80x150cm Motifs Ethniques Rouge Corail - - Meubles, Salons, Literie14 Jul 2023
BOLLY - Tapis Tissé 80x150cm Motifs Ethniques Rouge Corail - - Meubles, Salons, Literie14 Jul 2023- Tireuse à bière - PerfectDraft - Philips14 Jul 2023
 Tesla Model S/3/X/Y : Manette de jeu sans fil, Joystick - Torque Alliance14 Jul 2023
Tesla Model S/3/X/Y : Manette de jeu sans fil, Joystick - Torque Alliance14 Jul 2023 Des lunettes pour une meilleure vision nocturne au guidon. – Blog14 Jul 2023
Des lunettes pour une meilleure vision nocturne au guidon. – Blog14 Jul 2023 Le meilleur appareil photo compact en 2024 - Le guide complet14 Jul 2023
Le meilleur appareil photo compact en 2024 - Le guide complet14 Jul 2023 O que acontece com o corpo quando passamos a beber 8 copos d'água por dia? - 04/11/2016 - UOL VivaBem14 Jul 2023
O que acontece com o corpo quando passamos a beber 8 copos d'água por dia? - 04/11/2016 - UOL VivaBem14 Jul 2023 Prise socle tableau droit bleu 230V 2P+T 6H 32A IP55 Bals14 Jul 2023
Prise socle tableau droit bleu 230V 2P+T 6H 32A IP55 Bals14 Jul 2023

